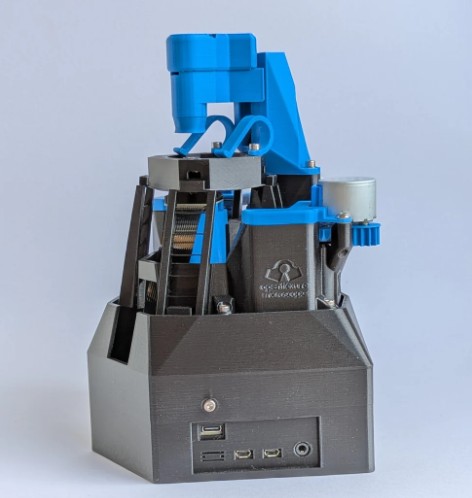
Brownian Motion with 3D-Printed Microscope
I built an openflexure microscope with printed parts, assembled the optics, and used Python to track Brownian motion of particles (micrometer scale). This page documents the build, the experiment, and my results.
Placeholder image — swap with a FINAL hero.